FLOGP
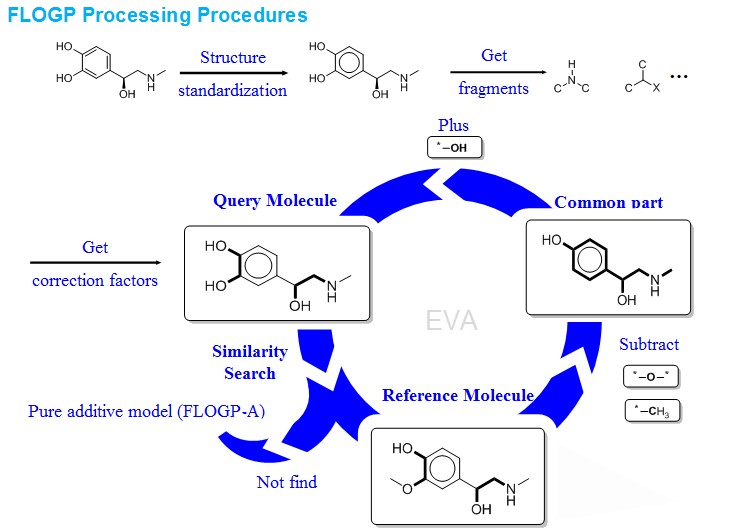
Fig. 1 The basic computational procedure of FLOGP.
The importance of the n-octanol/water partition coefficient (logP) as an indicator of lipophilicity in medical and environmental studies has been well established. Intra-molecular interaction has big influence on logP computation. To solve this problem, we proposed a new hybrid fingerprint (HFP) contained molecular fragment, intra-molecular interaction parameters and molecular descriptors and developed a new method, i.e., FLOGP, for logP computation. FLOGP predicts the logP value of a query compound by using the known logP value of a reference compound as a starting point. For a given query compound, the compound showing similar intra-molecular interaction in the reference database will be selected as the reference compound. It is calibrated on a training set of 15035 organic compounds with reliable logP data through partial least square (R = 0.962, RMSE = 0.501). FLOGP, as well as several popular logP methods, has been tested on a test set that includes 590 approved drugs. The result shows that our logP model can give a good prediction comparing with others (R = 0.945,RMSE=0.941).
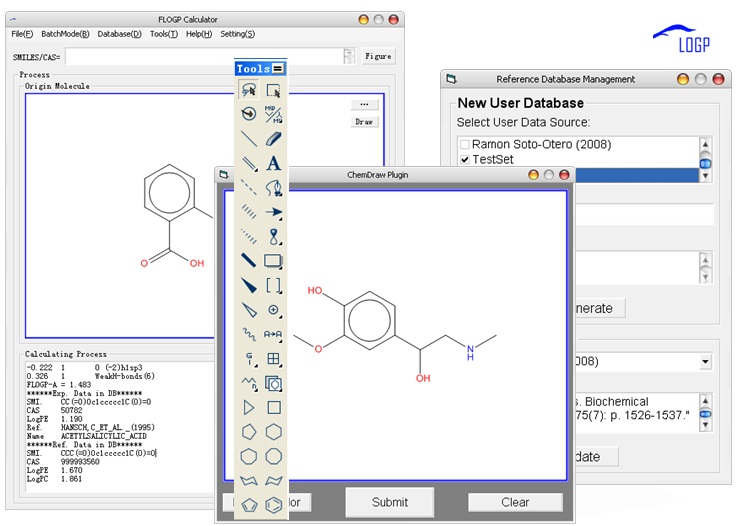
Fig. 2 Software screenshot of FLOGP
Download
Not available for download, but you can contact me for test and academic use. Because this work has not published yet.
GetBox-PyMOL-Plugin

Fig. 1 Screenshot of GetBox-PyMOL-Plugin
Autodock Vina and LeDock have been widely used in the simulation the interactions between proteins and small molecules. Docking Box is a key parameter for the docking. For the convenience and accuracy of the docking, this tool, Getbox-PyMOL-Plugin, is designed and created by me, which was first uploaded to BioMS forum in 2014, and uploaded to GitHub.
Docking box format
# Autodock Vina
--center_x xx.x --center_y xx.x --center_z xx.x --size_x xx.x --size_y xx.x --size_z xx.x
# LeDock
Binding pocket
xmin xmax
ymin ymax
zmin zmax
Download
available in github
Installation
Please follow the guiding steps shown in Fig 2. Open PyMOL->Plugin->Install Plugin->Find GetBox Plugin.py->Restart PyMOL->Finished, Then you will see an additional menu GetBox Plugin. it has three submenus. that is, Advanced usage, Autodetect box, Get box from selection (sele).

Fig. 2 Installation Procedures of GetBox Plugin
Usages
1. Autodetect box: Fetch the box with one click or use the code autobox 5.0
2. Get box from selection (sele) Select ligands or residues, then click the menu or use the code getbox (sele), 5.0
3. Advanced usages Using cmd mode with more flexibility
#e.g. autobox
#e.g. getbox
#e.g. getbox (sele), 6.0
#e.g. resibox resi 214+226+245, 8.0
#e.g. resibox resi 234 + resn HEM, 6.0
#e.g. showbox 2,3,4,5,6,7
Pocket Picker C

Fig. 1 Screenshot of Pocket Picker C
I rewrite the code of Pocket Picker in C language. The speed of C version is faster than previous python version. And a PyMOL Plugin is also designed to show the pocket that we find.
Download
Not available for download, but you can contact me for test and academic use.
Virtual Lib Generator

Fig. 1 Screenshot of Virtual Lib Generator
Here we developed a java version fragment decompose tool and virtual compounds library generator. The open-source CDK, RDKit and Indigochemoinformatics toolkits are combined in the program. CDK and RDKit Toolkit was used to decompose molecules into Murcko fragments and Ring systems. RDKit Toolkit was used to decompose molecules into fragment based on the rule of RECAP and BRICKS Indigo toolkits was used to generate a virtual compound library based on reaction rules.
Download
Not available for download, but you can contact me for test and academic use.
Docking Manager

Fig. 1 Screenshot of Docking Manager
To simplify the procedures of drug virtual screening, and manage the result data effectively, Docking Manager is developed to deal with this effectively. The software is based on Autodock vina, Open Babel and PyMol. The software also supports dividing jobs to 2 or more computers.
image2mol

Fig. 1 Screenshot of image2mol
In order to solve the problem of recognizing and translating chemical structures in image documents, a lot of software have been developed (ChemOCR, Imago, OSRA, Kekulé, CLiDE, ChemReader). But their GUI interface is not good for use, especially not suitable for chemical database construction. To accelerate the speed of chemical data collection for our NAIs database, A GUI for imago, image2mol, is developed. Like screen capture software, image2mol is very easy to use. You can capture the chemical structures in the papers or other places on your computer screen, and then edits search online or native databases.
Download
Not available for download, but you can contact me for test and academic use.